
Tutorial: Manipulating SpatialMap Objects
Tutorial_Manipulating_SpatialMap_objects.Rmd
library(facil)
library(SpatialMap)Basic operations
Subset, merge, and group
To get started quickly, let’s load a toy SpatialMap (sm_tonsil) object containing CODEX data.
sm_tonsil <- load_sm_data("tonsil")SpatialMap (SM) provides some simple functions to make it easier to cut your data or conduct grouped analyses.
Subset
Users may want to subset SM objects by removing regions from their analysis. They might also want to subset their object based on cells or cell metadata, e.g. if they want to pull out only one or more cell classes for further analysis. Both are easily accomplished with somewhat different syntaxes. By Regions:
## Keep only specific Regions.
## You can index Regions numerically or by name.
## This works just like base R.
sm2 <- sm_tonsil[2]
sm2
#> SpatialMap object
#> Active analysis: regions
#> + 10,694 total cells
#> + 16 features
#> + 1 Regions:
#> - TonsilB
sm2 <- sm_tonsil["TonsilA"]
sm2
#> SpatialMap object
#> Active analysis: regions
#> + 5,810 total cells
#> + 16 features
#> + 1 Regions:
#> - TonsilAAnd by cell annotations: (note: for the purposes of this demo, I am saving subset outputs to a new SM object, but that isn’t strictly necessary.)
## First, let's make some toy annotations
toy <- sample(letters, size = length(cells(sm_tonsil)), replace = T)
sm_tonsil <- addCellMetadata(sm_tonsil, toy, col.names = "some_letters")
## Now subset by the cell annotations
sm2 <- smSubset(sm_tonsil, on = "some_letters", keep = letters[1:10])
head(cellMetadata(sm_tonsil)) ## Note we only see the first part of the alphabet in the metadata now
#> cell_id cell.id anno316_some_categories anno916_Leiden_v1
#> TonsilA.125182 125182 TonsilA.125182 yay Cluster 2
#> TonsilA.125238 125238 TonsilA.125238 nay Cluster 2
#> TonsilA.125254 125254 TonsilA.125254 yay Cluster 1
#> TonsilA.125325 125325 TonsilA.125325 yay Cluster 1
#> TonsilA.125326 125326 TonsilA.125326 nay Cluster 6
#> TonsilA.125608 125608 TonsilA.125608 nay Cluster 1
#> anno248_test_subset_annotation_324
#> TonsilA.125182 p
#> TonsilA.125238 u
#> TonsilA.125254 t
#> TonsilA.125325 t
#> TonsilA.125326 b
#> TonsilA.125608 y
#> anno1189_uqc_test10_letters_qc_assoc anno1149_uqc_test10_letters
#> TonsilA.125182 b b
#> TonsilA.125238 b b
#> TonsilA.125254 b b
#> TonsilA.125325 c c
#> TonsilA.125326 c c
#> TonsilA.125608 b b
#> anno1133_uqc_test03_letters anno247_test_letters_324
#> TonsilA.125182 l p
#> TonsilA.125238 k u
#> TonsilA.125254 z t
#> TonsilA.125325 f t
#> TonsilA.125326 f b
#> TonsilA.125608 m y
#> anno1190_uqc_test10_letters_qc_assoc_postfilt
#> TonsilA.125182 <NA>
#> TonsilA.125238 <NA>
#> TonsilA.125254 b
#> TonsilA.125325 <NA>
#> TonsilA.125326 <NA>
#> TonsilA.125608 <NA>
#> anno253_Hierarchy.1 anno1797_uqc_test10_letters_221213
#> TonsilA.125182 not Th c
#> TonsilA.125238 not Th b
#> TonsilA.125254 Epithelial Cells b
#> TonsilA.125325 Epithelial Cells b
#> TonsilA.125326 Endothelial Cells b
#> TonsilA.125608 Epithelial Cells b
#> anno254_Neighborhood.clustering.based.on.annotation.set.253
#> TonsilA.125182 neighborhood_2
#> TonsilA.125238 neighborhood_2
#> TonsilA.125254 neighborhood_0
#> TonsilA.125325 neighborhood_0
#> TonsilA.125326 neighborhood_2
#> TonsilA.125608 neighborhood_0
#> anno1145_uqc_test08_letters anno1143_uqc_test08_letters Region
#> TonsilA.125182 l l TonsilA
#> TonsilA.125238 k k TonsilA
#> TonsilA.125254 z z TonsilA
#> TonsilA.125325 f f TonsilA
#> TonsilA.125326 f f TonsilA
#> TonsilA.125608 m m TonsilA
#> color width n_channels height
#> TonsilA.125182 grayscale 9408 4 9072
#> TonsilA.125238 grayscale 9408 4 9072
#> TonsilA.125254 grayscale 9408 4 9072
#> TonsilA.125325 grayscale 9408 4 9072
#> TonsilA.125326 grayscale 9408 4 9072
#> TonsilA.125608 grayscale 9408 4 9072
#> projection_visualizer_uuid sample_label n_levels
#> TonsilA.125182 96e6af5a-f612-4f97-a294-c580dcd41915 EnableShare 5
#> TonsilA.125238 96e6af5a-f612-4f97-a294-c580dcd41915 EnableShare 5
#> TonsilA.125254 96e6af5a-f612-4f97-a294-c580dcd41915 EnableShare 5
#> TonsilA.125325 96e6af5a-f612-4f97-a294-c580dcd41915 EnableShare 5
#> TonsilA.125326 96e6af5a-f612-4f97-a294-c580dcd41915 EnableShare 5
#> TonsilA.125608 96e6af5a-f612-4f97-a294-c580dcd41915 EnableShare 5
#> assay_id n_cycles base_image_visualizer_uuid
#> TonsilA.125182 1 8 728d42b7-77ea-4319-a48a-7f7340bf182d
#> TonsilA.125238 1 8 728d42b7-77ea-4319-a48a-7f7340bf182d
#> TonsilA.125254 1 8 728d42b7-77ea-4319-a48a-7f7340bf182d
#> TonsilA.125325 1 8 728d42b7-77ea-4319-a48a-7f7340bf182d
#> TonsilA.125326 1 8 728d42b7-77ea-4319-a48a-7f7340bf182d
#> TonsilA.125608 1 8 728d42b7-77ea-4319-a48a-7f7340bf182d
#> experiment_label segmentation_visualizer_uuid axes
#> TonsilA.125182 enableshare 9d6258e3-1532-40fb-9503-0831f8661358 TCYX
#> TonsilA.125238 enableshare 9d6258e3-1532-40fb-9503-0831f8661358 TCYX
#> TonsilA.125254 enableshare 9d6258e3-1532-40fb-9503-0831f8661358 TCYX
#> TonsilA.125325 enableshare 9d6258e3-1532-40fb-9503-0831f8661358 TCYX
#> TonsilA.125326 enableshare 9d6258e3-1532-40fb-9503-0831f8661358 TCYX
#> TonsilA.125608 enableshare 9d6258e3-1532-40fb-9503-0831f8661358 TCYX
#> assay_metadata assay_description
#> TonsilA.125182 {"image_mpp": 0.377, "num_amp_cycles": 0}
#> TonsilA.125238 {"image_mpp": 0.377, "num_amp_cycles": 0}
#> TonsilA.125254 {"image_mpp": 0.377, "num_amp_cycles": 0}
#> TonsilA.125325 {"image_mpp": 0.377, "num_amp_cycles": 0}
#> TonsilA.125326 {"image_mpp": 0.377, "num_amp_cycles": 0}
#> TonsilA.125608 {"image_mpp": 0.377, "num_amp_cycles": 0}
#> experiment_id visual_quality assay_name region_display_label
#> TonsilA.125182 164 true CODEX EnableShare
#> TonsilA.125238 164 true CODEX EnableShare
#> TonsilA.125254 164 true CODEX EnableShare
#> TonsilA.125325 164 true CODEX EnableShare
#> TonsilA.125326 164 true CODEX EnableShare
#> TonsilA.125608 164 true CODEX EnableShare
#> biomarker_expression_version segmentation_version study_name
#> TonsilA.125182 1 1 Enable Tonsil
#> TonsilA.125238 1 1 Enable Tonsil
#> TonsilA.125254 1 1 Enable Tonsil
#> TonsilA.125325 1 1 Enable Tonsil
#> TonsilA.125326 1 1 Enable Tonsil
#> TonsilA.125608 1 1 Enable Tonsil
#> some_letters
#> TonsilA.125182 d
#> TonsilA.125238 q
#> TonsilA.125254 i
#> TonsilA.125325 r
#> TonsilA.125326 o
#> TonsilA.125608 b
## This also works if your cell-wise subset eliminates one or more Regions
## We can make more toy data, this time split by sample, to illustrate
toy1 <- sample(letters[1:13], size = dim(sm_tonsil[[1]])[1], replace = T)
toy2 <- sample(letters[14:26], size = dim(sm_tonsil[[2]])[1], replace = T)
sm_tonsil <- addCellMetadata(sm_tonsil, c(toy1, toy2), col.names = "other_letters")
sm2 <- smSubset(sm_tonsil, on = "other_letters", keep = letters[1:10])You can also subset by spatial coordinates using a special helper function. In the future, this will be extended to provide an interface for rapid spatial tiling of an image. The i and j parameters are interpreted as follows: * If i is a scalar, coordinates less than or equal to i are included * If i is a 2-length vector, the elements will be treated as min and max of the subset window, respectively * If i is a range, any coordinates within the range will be included, including any not named.
An example is below:
Merge
We can also readily combine SM objects:
smc1 <- sm_tonsil[1]
smc2 <- sm_tonsil[2]
smc <- combineMaps(list(smc1, smc2), preserve.Analyses = T)
#> - Note: Setting `regions` as activeAnalysis
#> - Note: Settings from the 1st SpatialMap object will be used after mergeNote: preserve.Analyses will preserve any grouped analyses. They will be removed if FALSE.
Group
Finally, a powerful way to use SM objects is to conduct analyses across groups of samples. For example, you might be interested in conducting a normalization step for a 10-batch, 300-sample study, where normalization factors are calculated across batch Or you might be interested in using single cell genomics-style integration techniques, but want to group Regions by experiment.
These workflows are flexibly implemented by passing any projectMetadata column name to the analyze parameter of SpatialMap functions.
sm_tonsil <- Normalize(sm_tonsil, method = "robust", analyze = "study_name")
#> Normalizing data using method `robust`
#>
## The resulting medians from robust scaling are the same for markers across both samples
featureInfo(sm_tonsil)
#> $TonsilA
#> features robust_scale_medians robust_scale_mads
#> CD3e CD3e 139.4040 0.50585231
#> CD21 CD21 732.0685 1.97025621
#> CD68 CD68 283.5075 0.49878480
#> Actin Actin 93.0705 0.12765397
#> CD45RO CD45RO 637.7615 1.66434967
#> HisH3p HisH3p 118.1990 0.26227071
#> PanCK PanCK 33.5070 0.02573614
#> Ki67 Ki67 796.8080 3.09614671
#> CD11c CD11c 148.0615 0.15076952
#> ECad ECad 410.4025 1.03434516
#> CD4 CD4 168.8435 0.51752758
#> CD31 CD31 87.4295 0.19883195
#> Podoplanin Podoplanin 425.9225 1.28857259
#> CD107a CD107a 374.2065 0.71060740
#> CD20 CD20 892.5940 2.93562257
#> CD8 CD8 333.3275 1.01267271
#>
#> $TonsilB
#> features robust_scale_medians robust_scale_mads
#> CD3e CD3e 139.4040 0.50585231
#> CD21 CD21 732.0685 1.97025621
#> CD68 CD68 283.5075 0.49878480
#> Actin Actin 93.0705 0.12765397
#> CD45RO CD45RO 637.7615 1.66434967
#> HisH3p HisH3p 118.1990 0.26227071
#> PanCK PanCK 33.5070 0.02573614
#> Ki67 Ki67 796.8080 3.09614671
#> CD11c CD11c 148.0615 0.15076952
#> ECad ECad 410.4025 1.03434516
#> CD4 CD4 168.8435 0.51752758
#> CD31 CD31 87.4295 0.19883195
#> Podoplanin Podoplanin 425.9225 1.28857259
#> CD107a CD107a 374.2065 0.71060740
#> CD20 CD20 892.5940 2.93562257
#> CD8 CD8 333.3275 1.01267271Formal “Analyses”
Persistent groupings
We also might want to instantiate a more permanent grouping. For example, to analyze all SM object cells as one group, regardless of Region, we may want to perform dimensionality reduction and clustering on all cells.
In this case, the user can create an Analysis:
sm_tonsil <- createAnalysis(sm_tonsil)
#> New analysis created.
#> 2 Regions
#> 16504 cells
#> 16 common features
#>
#> Added Analysis `combined` to SpatialMap project 'SpatialMap Project'.
#> Setting as active Analysis
#> By default, an analysis called "combined" will be made, which includes all Regions in the SM object. These defaults can be changed with regions and id parameters. Feature subsets can also be passed into this function via the features parameter in createAnalysis.
Now, any SM function will automatically apply across all cells and their data at once.
sm_tonsil <- sm_tonsil %>%
Normalize(method = "scale", from = "NormalizedData", to = "ScaledData") %>%
runPCA(dims = 15)
#> Normalizing data using method `scale`
#>
#> By analysis: 'combined'
#>
#> By analysis: 'combined'
#>
A <- getAnalysis(sm_tonsil, "combined")
Representations(A, "pca")
#> Representation object 'pca'
#> Dimensions: 'PC1', 'PC2', 'PC3', 'PC4', 'PC5', 'PC6', 'PC7', 'PC8', 'PC9', 'PC10', 'PC11', 'PC12', 'PC13', 'PC14', 'PC15'
#> 16504 cellsNote that all SpatialMap functions should work like normal in this framework!
sm2 <- smSubset(sm_tonsil, on = "other_letters", keep = letters[1:10])
#> Warning in FUN(X[[i]], ...): Dropping Regions and cells from existing Analyses
#> may interfere with Representations and nearest neighbor networks.
smc1 <- createAnalysis(smc1)
#> New analysis created.
#> 1 Regions
#> 5810 cells
#> 16 common features
#>
#> Added Analysis `combined` to SpatialMap project 'SpatialMap Project'.
#> Setting as active Analysis
#>
combineMaps(list(smc1, smc2))
#> - Note: Setting `regions` as activeAnalysis
#> - Note: Settings from the 1st SpatialMap object will be used after merge
#> SpatialMap object
#> Active analysis: regions
#> + 16,504 total cells
#> + 16 features
#> + 2 Regions:
#> - TonsilA
#> - TonsilB
sm_tonsil[1]
#> Warning in FUN(X[[i]], ...): Dropping Regions and cells from existing Analyses
#> may interfere with Representations and nearest neighbor networks.
#> SpatialMap object
#> Active analysis: combined
#> + 5,810 total cells
#> + 16 features
#> + 1 Regions:
#> - TonsilAUnderstanding activeAnalysis
When creating an Analysis, you also see that the activeAnalysis is set to the new combined one. To revert to the default per-Region iteration behavior of SM functions, you can reset this to “regions”:
activeAnalysis(sm_tonsil) <- "regions"
sm_tonsil <- Normalize(sm_tonsil, method = "robust")
#> Normalizing data using method `robust`
#>
featureInfo(sm_tonsil)
#> $TonsilA
#> features robust_scale_medians robust_scale_mads
#> CD3e CD3e 49.6145 0.14748487
#> CD21 CD21 605.8785 2.67889453
#> CD68 CD68 259.7695 0.63240854
#> Actin Actin 45.6465 0.04786049
#> CD45RO CD45RO 242.2745 0.63040589
#> HisH3p HisH3p 70.6130 0.13973836
#> PanCK PanCK 64.8410 0.36256968
#> Ki67 Ki67 195.4330 0.75114192
#> CD11c CD11c 120.0590 0.24455030
#> ECad ECad 358.0260 1.70107349
#> CD4 CD4 96.2305 0.35832791
#> CD31 CD31 44.4345 0.08644003
#> Podoplanin Podoplanin 560.9920 4.19302742
#> CD107a CD107a 225.7370 0.85480405
#> CD20 CD20 612.6625 2.54358797
#> CD8 CD8 162.9525 0.62768455
#>
#> $TonsilB
#> features robust_scale_medians robust_scale_mads
#> CD3e CD3e 298.5845 0.83408283
#> CD21 CD21 814.6300 1.92029195
#> CD68 CD68 300.7225 0.48213692
#> Actin Actin 105.1175 0.04722075
#> CD45RO CD45RO 832.5930 0.94407279
#> HisH3p HisH3p 171.7610 0.27555744
#> PanCK PanCK 31.5755 0.01342539
#> Ki67 Ki67 1169.2360 2.82118632
#> CD11c CD11c 158.1795 0.10152008
#> ECad ECad 439.6465 0.85139381
#> CD4 CD4 265.4335 0.71193507
#> CD31 CD31 109.5900 0.13456306
#> Podoplanin Podoplanin 405.0540 0.79968561
#> CD107a CD107a 466.0280 0.56079488
#> CD20 CD20 1426.5280 4.31479541
#> CD8 CD8 505.5175 1.18733769The analyze parameter
You can also override the active Analysis temporarily by explicitly setting the analyze parameter at any time.
sm_tonsil <- Normalize(sm_tonsil, method = "robust", analyze = "combined")
#> Normalizing data using method `robust`
#>
#> By analysis: 'combined'
#>
featureInfo(sm_tonsil)
#> $TonsilA
#> features robust_scale_medians robust_scale_mads
#> CD3e CD3e 139.4040 0.50585231
#> CD21 CD21 732.0685 1.97025621
#> CD68 CD68 283.5075 0.49878480
#> Actin Actin 93.0705 0.12765397
#> CD45RO CD45RO 637.7615 1.66434967
#> HisH3p HisH3p 118.1990 0.26227071
#> PanCK PanCK 33.5070 0.02573614
#> Ki67 Ki67 796.8080 3.09614671
#> CD11c CD11c 148.0615 0.15076952
#> ECad ECad 410.4025 1.03434516
#> CD4 CD4 168.8435 0.51752758
#> CD31 CD31 87.4295 0.19883195
#> Podoplanin Podoplanin 425.9225 1.28857259
#> CD107a CD107a 374.2065 0.71060740
#> CD20 CD20 892.5940 2.93562257
#> CD8 CD8 333.3275 1.01267271
#>
#> $TonsilB
#> features robust_scale_medians robust_scale_mads
#> CD3e CD3e 139.4040 0.50585231
#> CD21 CD21 732.0685 1.97025621
#> CD68 CD68 283.5075 0.49878480
#> Actin Actin 93.0705 0.12765397
#> CD45RO CD45RO 637.7615 1.66434967
#> HisH3p HisH3p 118.1990 0.26227071
#> PanCK PanCK 33.5070 0.02573614
#> Ki67 Ki67 796.8080 3.09614671
#> CD11c CD11c 148.0615 0.15076952
#> ECad ECad 410.4025 1.03434516
#> CD4 CD4 168.8435 0.51752758
#> CD31 CD31 87.4295 0.19883195
#> Podoplanin Podoplanin 425.9225 1.28857259
#> CD107a CD107a 374.2065 0.71060740
#> CD20 CD20 892.5940 2.93562257
#> CD8 CD8 333.3275 1.01267271This is super useful when you don’t want to manually set activeAnalysis() over and over again, but want to check something on a Region level quickly.
For example, when working with a combined Analysis, spatial embeddings (xy coordinates) have no consistent meaning. Trying to plot a spatial Representation from a combined Analysis gives an error, but you can easily return a list of Region-spatial plots by explicitly setting the analyze parameter:
plotRepresentation(sm_tonsil, "spatial", "cd4", analyze = "regions")
#> $TonsilA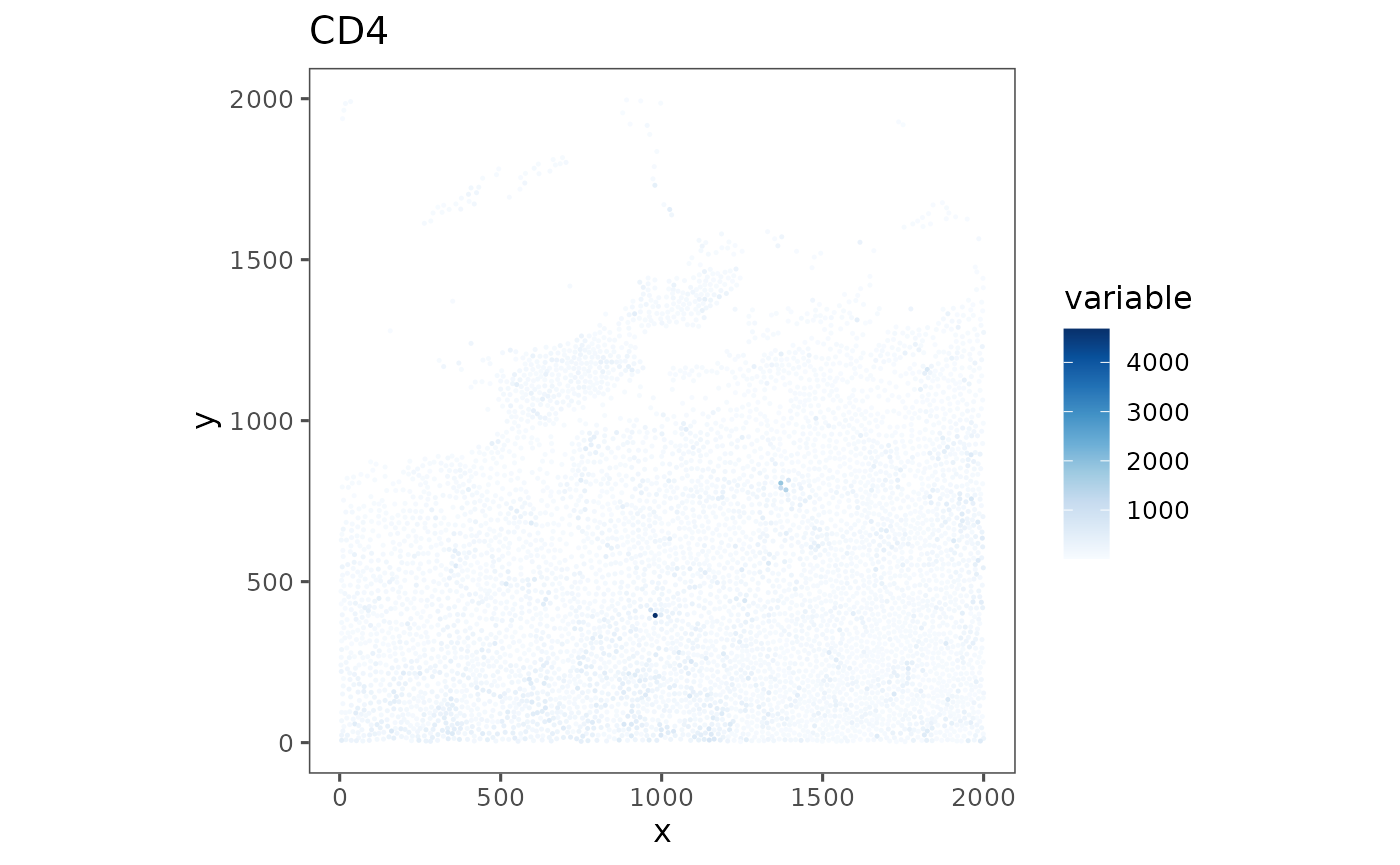
#>
#> $TonsilB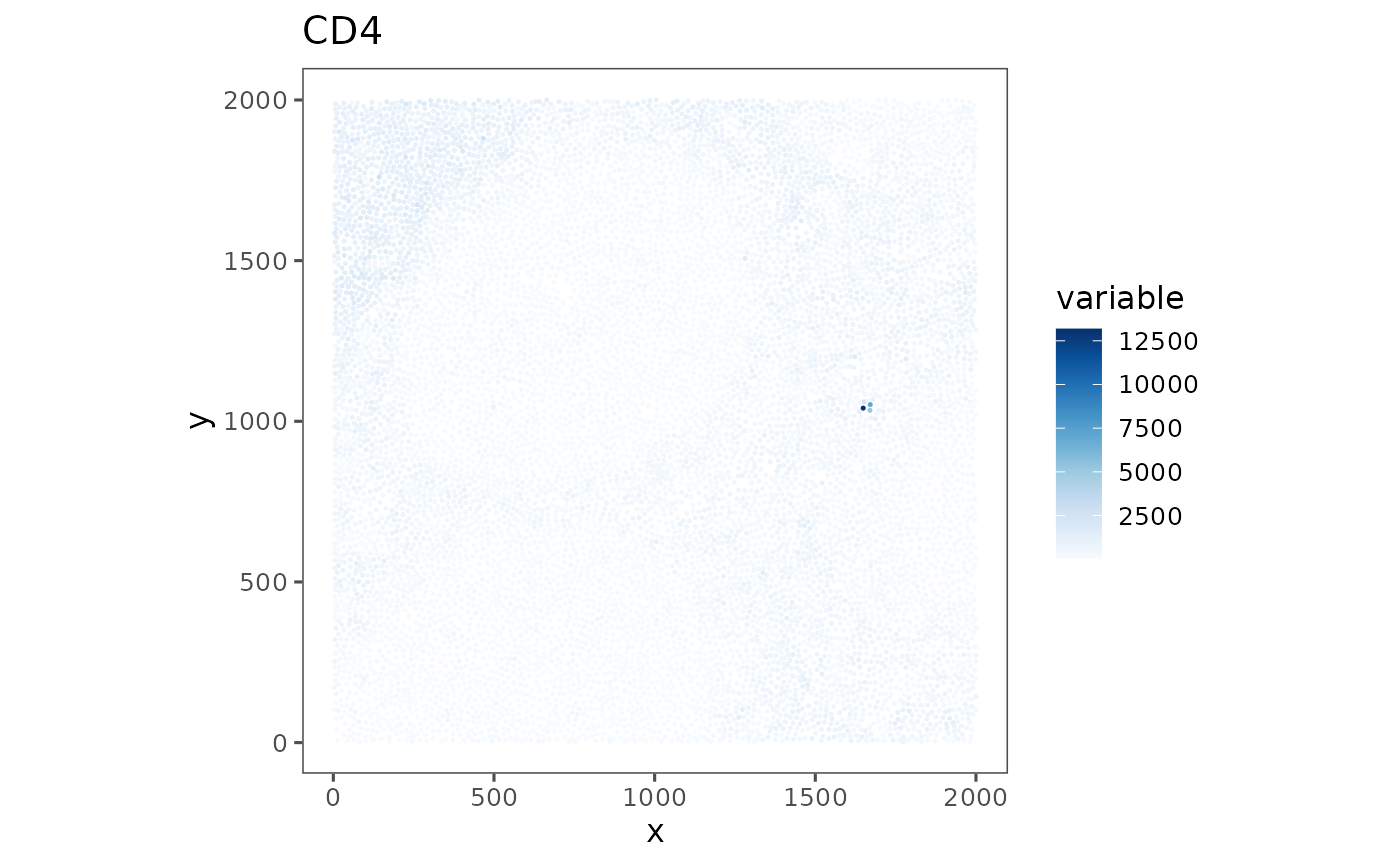
Finally, analyze is a useful debugging tool for the package. Much of SpatialMap’s complexity comes from the grouping structures and the flexibility built in there. Setting analyze = "regions" lets the package functions work in their most simple form.
Review
To review:
-
You can pass
-
"regions"(a special word in SM)
-
- a metadata grouping (e.g., “batch”), or
- an Analysis name (e.g.
"combined") to theanalyzeparameter of most SM functions! This built in flexibility makes it easier to switch modes between single-cell analyses, where spatial coordinates have no consistent meaning, and spatial analyses, where they do.
- an Analysis name (e.g.
Analyses are formal, persistent groupings that allow you to store results in a separate compartment from the original Regions.